King Phillip Came Over From Great Spain
A Classic of Biology Needs Some Serious Updating
I happened to hear a classic old tune I like the other day. If you guessed, 1994s breakout hit Regulate by the legendary Warren G, you’d be wrong. It was actually that stupid song for teaching kids about the Linnaeus classification system in biology. You know what I’m talking about right? the kingdom, phylum, class, order thing…..King Phillip came over from great Spain. If you ever had to take a science class in grades 2–8, especially if there was any sort of focus on biology it probably would have been forced on you at some point. The song is super annoying but kind of catchy. Lyrics and link if you are a glutton for punishment.
Kingdom phylum class and order, family genus species
All those plants and mammals, all the birds and fishies
Everything that flies and walks and all the things on leashes
Are kingdom phylum class and order, family genus species
Now classifying all of life’s a complicated game
And who made up the rules for this? Linnaeus was his name
He said we need a two-part name for everything in nature
The genus and the species that’s binomial nomenclature
(chorus)
It’s sort of like the countries, states, cities, neighborhoods
Each step down there’s fewer things to count, and that is good!
Through similarity of forms the groups were all created
But DNA’s our perfect clue to how we’re all related!
(chorus)
Our kingdom is animialia, our phylum is chordata
Mammalia’s that class of ours, the order is primata
Family’s hominidae, we’re getting near the end now
Homo sapiens is what we call you and your friends now!
(chorus)
Now the kingdoms got confusing so they added the domains
Archaea, Bacteria, Eukaryota were their names
Eukaryotes are most of the familiar life we’ve charted
But don’t forget that single cells are how all life got started!
(chorus)
The problem here is that the song ends at species. Yes, there is a sop to the domains which are a relatively recent addition but dude, come on? species? Thanks to advances in molecular biology we have come a very long way past the crude morphological/phenotypic (essentially appearance or the things we can observe about an organism) based classification system that Linnaeus developed. And we have now gone way beyond species to much greater levels of resolution in classifying organisms by how much alike and different they are. In the modern system these classifications are made by genetic sequence alignments. Things that meet a certain criteria, or threshhold of sameness to coin a phrase, in DNA sequence in specific parts of the genome are considered the same, while those that do not are considered different. One way this can be visualized is in the form of a neighbor joining tree which is a abstract representation of sameness based on genetic sequence data. I have included an example as an image below.
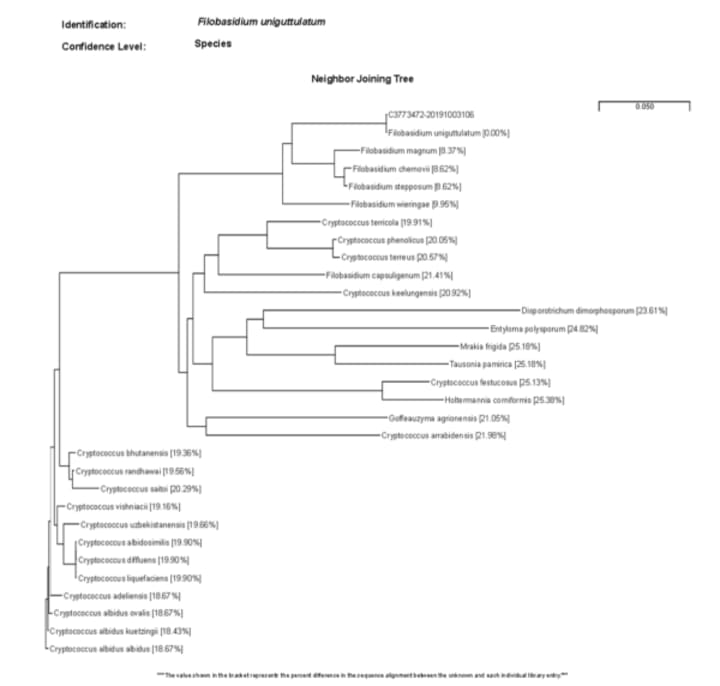
Just like with macro biological classification, micro and molecular biological classification allows for organisms to be the same at higher levels of classification, but differences become apparent as you go deeper into the levels. In other words, as you have more and more DNA sequence data available to use as a comparison. That glosses over a ton as it isn’t just about the availability of the data but also how you get that data (what technology is used) and how you process it (bioinformatics) but that is way more complicated then you probably care to read about. And definitely way more complicated then I feel like writing about.
So we can now come back full circle (or maybe half circle?) to the Linnaeus system and its shortcomings. As I mentioned it ends at the species level but now we have the capability to go much deeper. For bacteria at least the classification/naming/identification system now goes (in order from furthest camera pan out to highest level zoom). Genus → species →subspecies →serotype →strain →genotype. There are some some other oddball things mixed in their for certain species of bacteria but they are mostly still around for historical reasons. Also, to add to the confusion, some people use the terms serotype and strain interchangeably though they are not. To digress slightly, typically bacteria are referred to by the Genus and species to which the belong. For example Escherichia (Genus) coli (species). I say typically because there are some big exceptions, including a close cousin of E. coli, Salmonella. They are most often named Genus (Salmonella) followed by serotype (Typhimurium). Again this is mostly for historical reasons. Salmonella is a very interesting case because there are actually only two species of Salmonella, enterica and bongori, and six subspecies. However there are currently over 2700 serotypes. How were the serotypes determined you might be asking yourself? Now that is a long story, but suffice to say it was a long time ago, and it was not done with DNA sequencing approaches, which has resulted in no end of headaches for yours truly for the past few years. I have linked the WHO guidebook which lists all the serotypes for those who are interested below. It also defines precisely how they are to be written (I am only mostly following those guidelines in this article) which includes strict rules on the use of italics and capital/lower case letters etc. I use this reference almost everyday and it makes for an interesting if cryptic read for those who are not familiar with the field.
Identifying bacteria to the different classification levels requires the use of different techniques. Today these are mostly sequencing based though there are some notable exceptions. I will end with a note on genotype, the deepest one can go down the DNA sequence rabbit hole. Each unique living thing could be considered its own genotype as each unique living thing has some (very tiny in some cases) differences in their DNA sequences. Microbial clones are of course not unique individuals and thus all clones would be of the same genotype and share the exact same genetic makeup. They are also pretty sweet. In contrast to those other clones, from that reprehensible movie that crushed my hopes and dreams for two years that the Star Wars prequels could recover and still be good, Attack of the Clones. Uggh. I think I just threw up in my mouth a little. I will never forgive George Lucas for that….never.
About the Creator
Everyday Junglist
Practicing mage of the natural sciences (Ph.D. micro/mol bio), Thought middle manager, Everyday Junglist, Boulderer, Cat lover, No tie shoelace user, Humorist, Argan oil aficionado. Occasional LinkedIn & Facebook user






Comments (1)
This was cool to read! I'm excited for my microbiology class that it'll be a while until I can take.