Introduction of DNA Modifying Enzymes
DNA stores the genetic information on which organisms depend for survival and reproduction. The integrity of DNA molecules is critical to cellular function. So the DNA of human organism cells is constantly damaged by various external factors (such as ultraviolet light, electric radiation, chemical poisons, Figure 1) and endogenous factors (such as free radicals produced by metabolic intermediates). Cells can occur 104 times damages in a day. If the damage cannot be repaired in time, it will lead to apoptosis, uncontrolled cell growth, genetic variation and malignant tumors. For example, excessive exposure of human skin to strong sunlight can easily contribute to skin cancer. Mammalian DNA repair methods include direct repair (DR), mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), and double strand break repair (DSBR) including homologous recombination (HR) and non-homologous end joining (NHEJ).
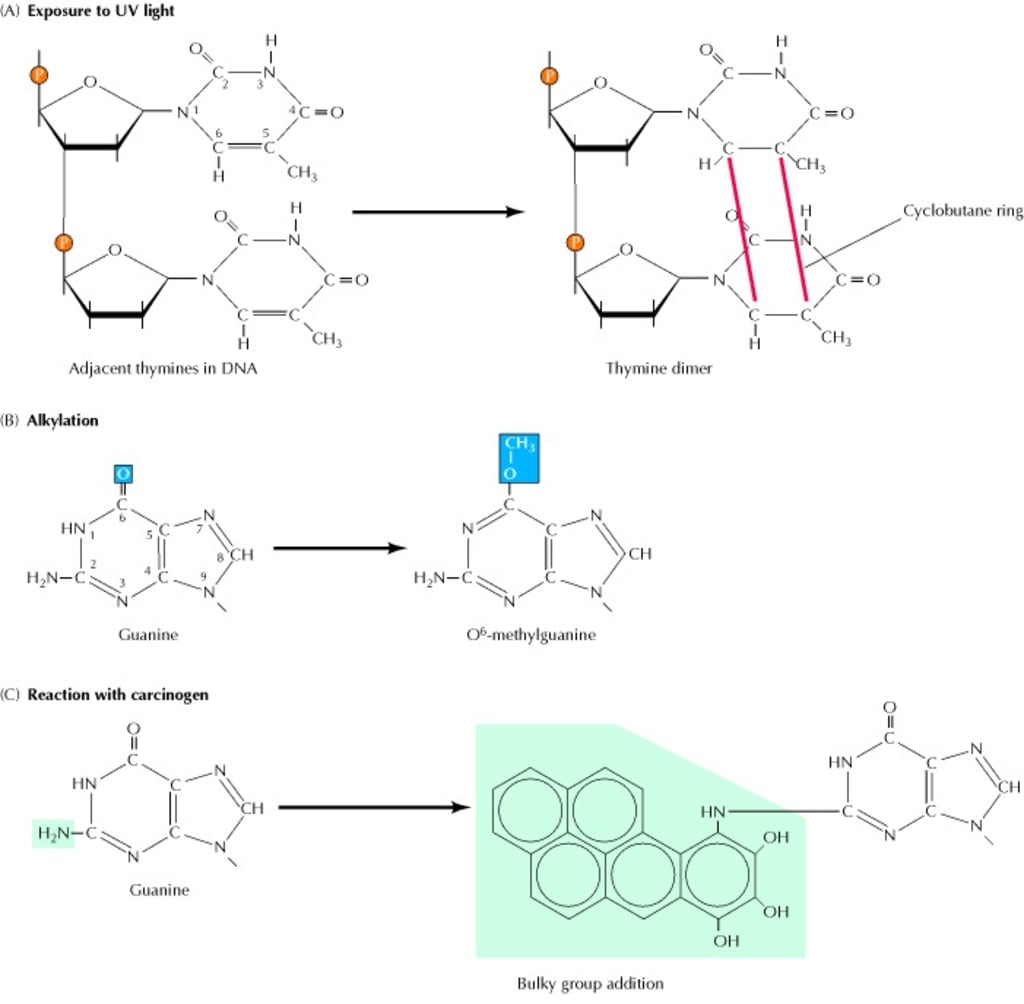
(A) UV light induces the formation of pyrimidine dimers, in which two adjacent pyrimidines (e.g., thymines) are joined by a cyclobutane ring structure. (B) Alkylation is the addition of methyl or ethyl groups to various positions on the DNA bases. In this example, alkylation of the O6 position of guanine results in formation of O6-methylguanine. (C) Many carcinogens (e.g., benzo-(a) pyrene) react with DNA bases, resulting in the addition of large bulky chemical groups to the DNA molecule.
Direct repair (DR)
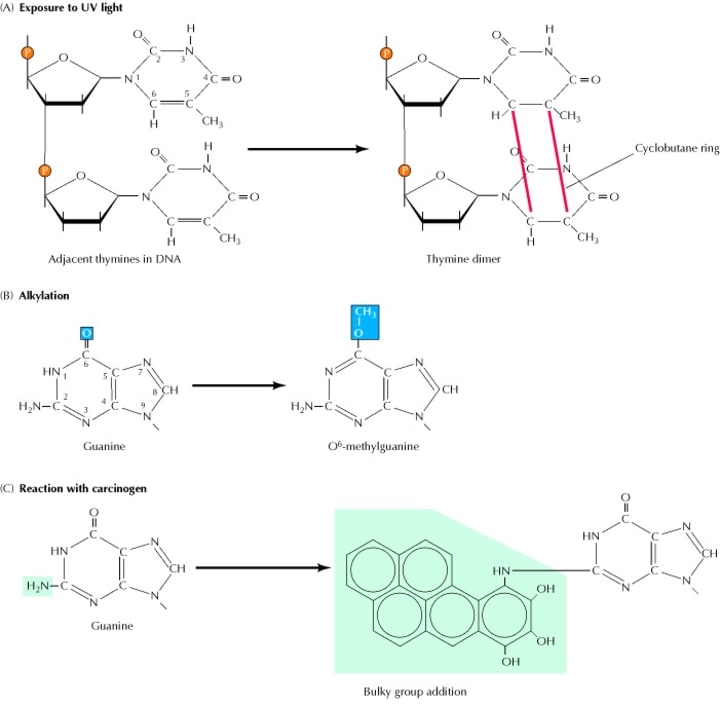
Direct repair is a simple way to repair DNA damage, and the damaged base can be recovered in one step. One important repair factor in DR is O6-alkylguanine DNA alkyltransferase (AGT), which is ubiquitous in the body. And the expression of this enzyme can be found in almost all types of cancer cells such as colon cancer cells, pancreatic cancer cells, and lung cancer cells. The resistance of tumor cells to the chlorinated drugs (carmustine, lomustine) and methylation drugs (temozolomide, TMZ) is closely related to the repair of AGT, among which O6-methylguanine DNA methyltransferase, MGMT, is the most important DR enzyme. When the alkylating agent acts on the DNA of the body cells, the methylation and alkylation damage of the guanine occurs at the O6 site. The MGMT can transfer the methyl group from the O6 position of O6-methylguanine to the cysteine residue at position 145 on itself, which will restore the guanine on the DNA strand and irreversibly inactivates itself (Figure 2). Therefore, MGMT can prevent DNA damage from alkylating groups and protect cells from tumors, but it also causes tumor cells to fight against alkylating agents and make tumor cells resistant to chemotherapeutic drugs.
Mismatch repair (MMR)
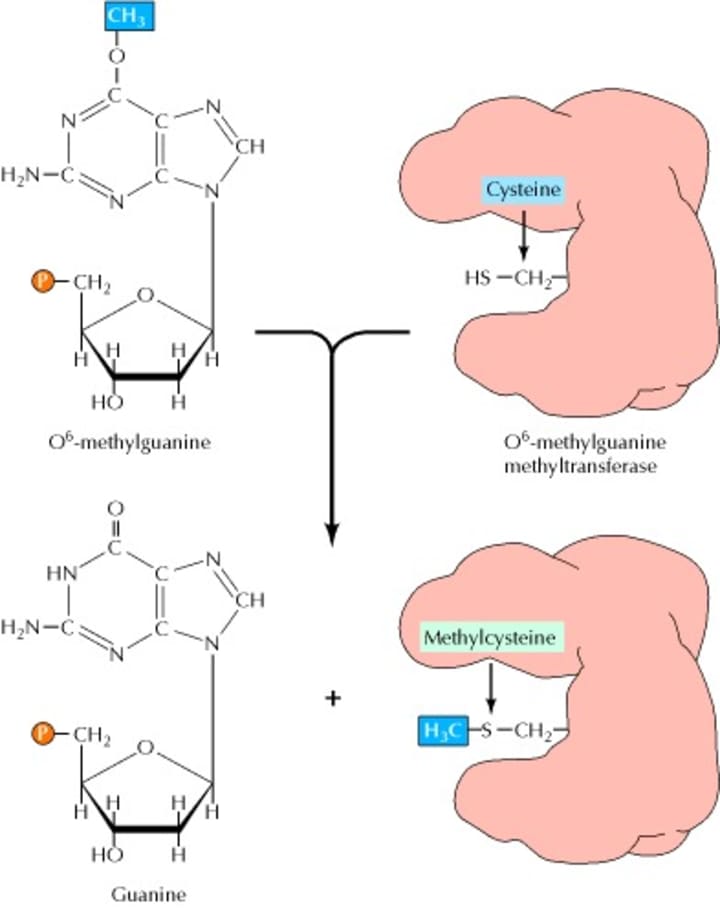
The mismatch repair system is mainly for the process of excision and repair of base mismatches generated during DNA recombination and replication. MMR in human cells mainly includes three processes of mismatch recognition, mismatch excision and DNA resynthesis (Figure 3), involving hMSH2, hMSH3, hMSH6, hMLH1, hMLH3, hPMS1 repair proteins. The DNA mismatch repair system recognizes and corrects mismatched DNA base pairs and plays a role in maintaining DNA replication fidelity and controlling gene variation. If the MMR function is defective, the repair of the mismatched base copy will fail, and then there will be an increase in the frequency of spontaneous mutation which causes instability of the genome and tumor production. The MMR system not only repairs errors in biosynthesis, but also participates in a variety of DNA damage-induced apoptosis processes. Defects in MMR function will directly affect the ability of cells to recognize DNA damage and activate apoptosis, making tumor cells resistant to some chemotherapeutic drugs. Studies have shown that MMR is closely related to the natural and acquired resistance of TMZ as well as cisplatin. Three of the short-segment repair enzymes in humans have been identified: T/G-specific thymidine DNA glycosylase, A/G-specific nickase, and DNA polymerase α which could recognize all types of mismatch. Their respective function is to remove the T in the T/G mismatch, to incise mismatched A on the 5' and 3' and to participate in DNA repair synthesis.
Base excision repair (BER)
BER is a multi-step complex system that requires several repair systems to participate in. BER plays an important role in maintaining the stability of the genome and inhibiting tumorigenesis. The repair process of BER is as follows: First, the DNA glycosidase excises the damaged base to form an apurinin/apyrimidimic site (AP site); then the AP endonuclease (apurinic/apyrimidinic endonuclease, APE) cuts the glycoside-phosphate bond of nucleotide and remove small fragment DNA including AP site nucleotides; DNA polymerase synthesizes new single nucleotides to complete alternative synthesis; the complex of DNA ligase I (or III) and XRCCI (X-ray repair cross- complementing 1) will close the interface and complete the repair (Figure 4). The main feature of BER is the release of unpaired bases of DNA catalyzed by DNA glycosylation enzymes. The enzymes and genes involved in BER repair mainly include XRCC1 (X-ray repair cross-complementing gene 1), DNA ligase, hOGG1 (8-oxoguanine DNA glycosylase 1), MPG (N-methylpurine DNA), APE (a-purinic endonuclease) and so on. XRCC1 which is sensitive to ionizing radiation is the first gene isolated from mammals and located at 19q13.2. Studies have shown that the use of antisense RNA technology to inhibit the endonuclease activity of APE1 can increase the sensitivity of pancreatic cancer cells to gemcitabine. It is also possible to improve the therapeutic effect of tumors by combining the application of BER inhibitors such as poly ADP-ribose polymerase (PARP) and APE1 inhibitors. For example, it has been reported that PARP inhibitors combined with TMZ can greatly enhance the anticancer activity of TMZ; another preclinical study has shown that APE1 inhibitors combined with alkylating agents can enhance the cytotoxic effects of temozolomide.
Nucleotide excision repair (NER)
NER is widely studied in bacteria, yeast, and human xeroderma pigmentosum (XP). The human NER system is very complex which can be divided into four phases: damage identification, incision, filling gaps and connections (Figure 5). The proteins involved in this process include XPA-G (xerodermap pigmentosum group A to G), replication protein A (RPA), replication factor C (RFC), PCNA (proliferating cell nuclear antigen), transcription factor TFIIH and so on.
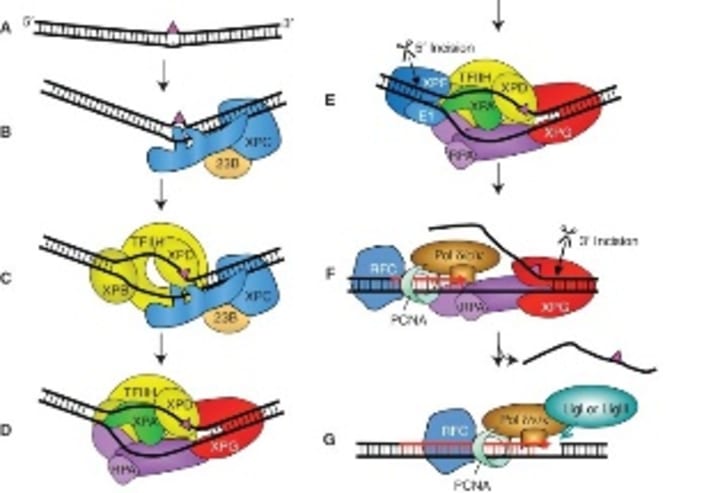
XPA to G are associated with human xeroderma pigmentosum. The complex of XPA and RPA is the damage recognition factor that determines the site of damage to the DNA; XPB forms a damage-recognition complex that plays a role in transcription-coupled repair. Dorota Butkiewicz et al have found that the polymorphism of XPD is closely related to the risk of non-small cell lung cancer. XPG has three nuclease activities: single-strand specific exonuclease activity, exonuclease activity at the 5' and 3' ends, and FEN (flap endonuclease) activity. XPF has a 5' endonuclease activity and may be involved in recombinant repair. The least known in NER is XPC and XPE. Masayuki Yokoid et al reported that the XPC-HR23B complex plays an important role in the early stages of genome-wide repair, especially in the identification of damage, and the formation of open complexes or repair protein complexes. The XPE may be another binding protein associated with UV-damaged DNA. The relationship between NER and human diseases is mainly the cancer susceptibility of XP patients. The risk of skin cancer in NER-deficient XP patients is about 1000 times higher than that of normal people.
Double strand break repair (DSBR)
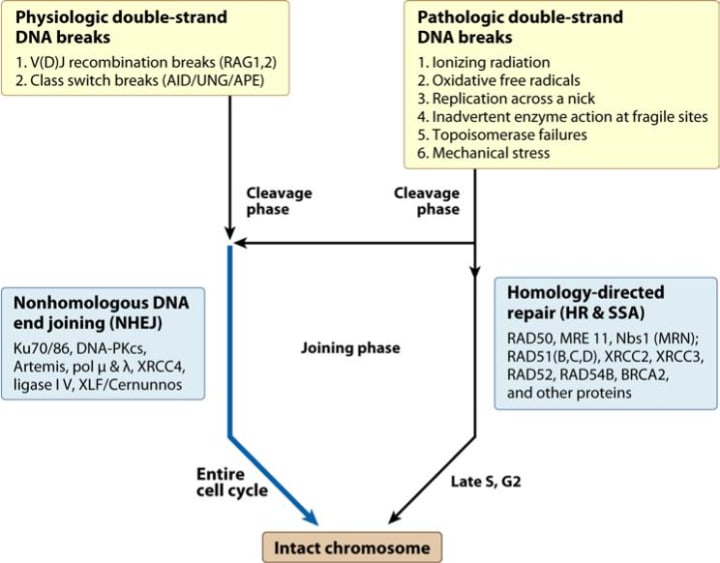
DNA double strand breaks (DSBs) can be caused by DNA metabolism, ionizing radiation, and reactive oxygen species damage. If DSBs cannot be repaired in time, DNA replication and transcription will be blocked. Therefore, effective repair of DSBs is essential for maintaining genome integrity and gene expression. DSBR includes two sub-pathways (Figure 6): homologous recombination (HR) and non-homologous end joining (NHEJ). Both pathways involve multiple repair elements, and through the complex process of multi-step reactions, the two pathways together maintain the stability of the cell genome. HR is a kind of precise repair, which synthesizes new DNA fragments by using the homologous sequence region of the complete DNA molecule as a template to maximize the accuracy of the genomic information. The preferred template is the sister chromatid of the injured molecule, and the repair process is generally in the late S to G2 phase. The mechanism of action can be summarized into three processes: first, processing of DNA damage sites; chain invasion and repair synthesis; formation and dissociation of Holliday intermediates. NHEJ is mediated by a DNA-dependent protein kinase that reconnecting broken DNA duplexes by direct action of DNA ligase. This pathway is usually accompanied by nucleotide insertion or deletion mutations and can be activated in various cycles of the cell. The DSBR mechanism is very complex and involves a large number of proteins such as Rad51, Rad52, Rad54, BRCA1, BRCA2, and p53. Rad51 protein is a key factor involved in DSBR. Studies have shown that increased expression of Rad51 protein will lead to enhanced DNA repair capacity, thereby increasing cell viability and ultimately leading to tumor cell resistance to chemotherapy. The most popular view now is that HR is the predominant form in yeast, and NHEJ is the predominant form in mammalian cells, but HR is also found in mammalian cells.
DNA repair inhibitors
With the deepening of research on the relationship between DNA repair and chemoresistance, how to reduce chemoresistance through DNA repair mechanism has gradually achieved some results. Among them, DNA repair inhibitors are a hot field in recent years, and they combined with chemotherapy drugs play an important role in anti-tumor. MGMT, PARP, and DNA-dependent protein kinase (DNA-PK) inhibitors have been used in clinical trials. In addition to MGMT, more research is on PARP inhibitors at present. PARP is a multifunctional protein which plays an important role in BER. Another DNA repair key factor, DNA-PK, belongs to the phosphoinositide 3-kinase (PI3K) family. It is a DNA-activated nuclear serine/threonine kinase that plays a key role in NHEJ. DNA-PK Inhibitors can greatly enhance the sensitivity of radiotherapy and also enhance the sensitivity of tumor cells to DNA damaging agents.
References:
- Cooper, GeoffreyM. The cell: a molecular approach /-2nd ed. ASM Press. 2010.
- Li G M. Mechanisms and functions of DNA mismatch repair. Cell Research. 2008, 18(1):85-98.
- Krokan H E, Bjørås M. Base excision repair. Cold Spring Harbor Perspectives in Biology. 2013, 5(4):a012583.
- Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harbor Perspectives in Biology. 2013, 5(10):a012609.
- Lieber M R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual Review of Biochemistry. 2010, 79(1):181-211.
About the Creator
Creative Diagnostics
Creative Diagnostics is a leading manufacturer and supplier of antibodies, viral antigens, innovative diagnostic components, and critical assay reagents.



Comments
Creative Diagnostics is not accepting comments at the moment
Want to show your support? Send them a one-off tip.